BIOTECHNOLOGY UNIT - 2 (cloning vector , restriction endonuclease, DNA ligase , recombinant DNA technology,application of genetic engineering , interferons , vaccines-hepatitis B , PCR )
Cloning vectors :-
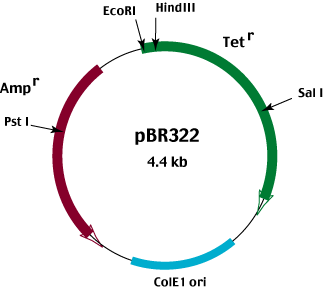
A cloning vector is a genome that can accept the target DNA and increase the number of copies through its own autonomous replication. It can be a plasmid, a bacteriophage, or yeast artificial chromosome (YAC). Cloning vectors usually are selected on the basis of differences in their capacity for the size of the insert DNA.
Many types of vector and host cells are employed in cloning experiments. The choice of vector depends on the size of the insert to be cloned and the purpose of the experiment .
Plasmids and lambda bacteriophage vectors are used for cloning small inserts of up to a few kilobases of DNA; cosmid vectors, on the other hand, can replicate efficiently with up to about 50kb of DNA insert; bacterial artificial chromosomes (BACs) and yeast artificial chromosomes (YACs) are capable of replicating significantly larger inserts of up to about 500kb of DNA (BACs) and more than 1000kb of DNA (YACs).
.Experience showed that whereas YACs were capable of holding more 1 megabase of insert DNA, they tended to become unstable, causing the inserts to rearrange, whereas BACs were found to be far more stable and have become the mainstay for many genome projects.
RECOMBINANT DNA TECHNOLOGY :-
Recombinant DNA is the general name for taking a piece of one DNA, and combining it with another strand of DNA.
Thus the name recombinant DNA is also referred as "chimera" .
By combining two or more different strands of DNA, scientists are able to create a new strand of DNA.
The most common recombinant process involves combining the DNA of two different organisms.
.Recombinant DNA formation process:-
There are three different methods by which Recombinant DNA is made. They are :-
1.Transformation,
2.Phage Introduction, and
3.Non-Bacterial Transformation.
Transformation
The first step in transformation is to select a piece of DNA to be inserted
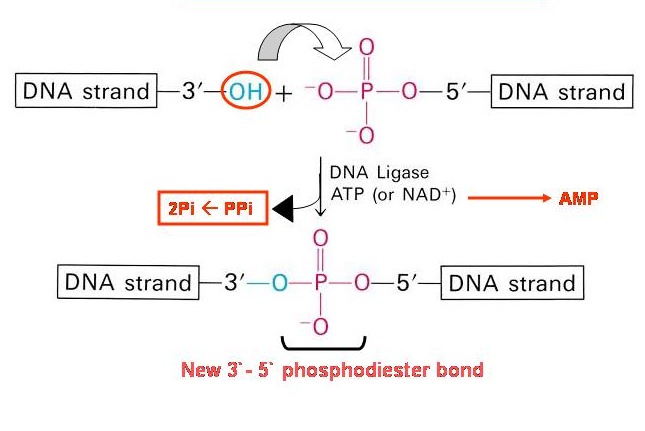
into a vector. The second step is to cut that piece of DNA with a restriction
enzyme and then ligate the DNA insert into the vector with DNA Ligase.
The insert contains a selectable marker which allows for identification of recombinant molecules.
An antibiotic marker is often used so a host cell without a vector dies when exposed to a certain antibiotic, and the host with the vector will live because it is resistant.
The vector is inserted into a host cell, in a process called transformation. One
example of a possible host cell is E. Coli. The host cells must be specially
prepared to take up the foreign DNA.
Selectable markers can be for antibiotic resistance, color changes, or any other
characteristic which can distinguish transformed hosts from untransformed hosts.
Different vectors have different properties to make them suitable to different
applications. Some properties can include symmetrical cloning sites, size, and
high copy number.
Non-Bacterial Transformation
This is a process very similar to Transformation, which was described above. The
only difference between the two is non-bacterial does not use bacteria such as E. Coli
for the host. Ex-microinjections,biolistics etc
In microinjection, the DNA is injected directly into the nucleus of the cell being
transformed.
In biolistics, the host cells are bombarded with high velocity microprojectiles, such as particles of gold or tungsten that have been coated with DNA.
Phage Introduction
Phage introduction is the process of transfection, which is equivalent to transformation,
except a phage is used instead of bacteria. In vitro packagings of a vector is used.
This uses lambda or MI3 phages to produce phage plaques which contain recombinants.
The recombinants that are created can be identified by differences in the
recombinants and non-recombinants using various selection methods.
Application of genetic engineering in medicines :-
Applications of rDNA (recombinant DNA) technology and genetic engineering in production of :-
1.Interferons:-
Interferons are a group of signaling proteins made and released by host cells in response to the presence of several viruses. In a typical scenario, a virus-infected cell will release interferons causing nearby cells to heighten their anti-viral defenses.
A wide variety of substances can induce the production of interferon, the most obvious of which is the animal viruses.
It is matter of research whether the virus contains DNA or RNA or if it is a live or killed virus.
They do, however, vary in their ability to stimulate interferon production with the paramyxoviruses (eg Newcastle disease virus, Sendai virus) and the arboviruses (eg Semliki Forest virus) being among the most efficient.
Bacteria, phages, chlamydiae, rickettsiae, mycoplasmas, protozoa and fungi are among the other, mainly intracellular organisms, that can stimulate interferon production.
The whole organism is not necessary for the induction of interferon production; fungal extracts, endotoxins, exotoxins and various lipid and sugar fractions such as capsular polysaccharide are also effective.
The genes controlling the production of interferon a and probably that of interferon f8 are situated on chromosome 9, and that for interferon y on chromosome 12.
2.Vaccines-Hepatitis B :-
In July 1986, a recombinant hepatitis B vaccine was licensed in the United States.
This vaccine built on the knowledge that heat-inactivated serum containing hepatitis B virus (HBV) and hepatitis B surface antigen (HBsAg) was not infectious, but was immunogenic and partially protective against subsequent exposure to HBV.
HBsAg was the component that conferred protection to HBV on immunization.To produce this vaccine, the gene coding for HBsAg, or “S” gene, was inserted into an expression vector that was capable of directing the synthesis of large quantities of HBsAg in Saccharomyces cerevisiae.
The HBsAg particles expressed by and purified from the yeast cells have been demonstrated to be equivalent to the HBsAg derived from the plasma of the blood of hepatitis B chronic carriers.
The recombinant S. cerevisiae cells expressing HBsAg are grown in stirred tank fermenters. The medium used in this process is a complex fermentation medium that consists of an extract of yeast, soy peptone, dextrose, amino acids, and mineral salts.
In-process testing is conducted on the fermentation product to determine the percentage of host cells with the expression construct. At the end of the fermentation process, the HBsAg is harvested by lysing the yeast cells. It is separated by hydrophobic interaction and size-exclusion chromatography. The resulting HBsAg is assembled into 22-nm–diameter lipoprotein particles. The HBsAg is purified to greater than 99% for protein by a series of physical and chemical methods. The purified protein is treated in phosphate buffer with formaldehyde, sterile filtered, and then coprecipitated with alum (potassium aluminum sulfate) to form bulk vaccine adjuvanted with amorphous aluminum hydroxyphosphate sulfate. The vaccine contains no detectable yeast DNA but may contain not more than 1% yeast protein. In a second recombinant hepatitis B vaccine, the surface antigen expressed in S. cerevisiae cells is purified by several physiochemical steps and formulated as a suspension of the antigen absorbed on aluminum hydroxide.
The procedures used in its manufacturing result in a product that contains no more than 5% yeast protein.
No substances of human origin are used in its manufacture.
Vaccines against hepatitis B prepared from recombinant yeast cultures are noninfectious and are free of association with human blood and blood products.
Each lot of hepatitis B vaccine is tested for safety, in mice and guinea pigs, and for sterility.
QC product testing for purity and identity includes numerous chemical, biochemical, and physical assays on the final product to assure thorough characterization and lot-to-lot consistency. Quantitative immunoassays using monoclonal antibodies can be used to measure the presence of high levels of key epitopes on the yeast-derived HBsAg.
A mouse potency assay is also used to measure the immunogenicity of hepatitis B vaccines.
The effective dose capable of seroconverting 50% of the mice (ED50) is calculated.
Hepatitis B vaccines are sterile suspensions for intramuscular injection. The vaccine is supplied in four formulations: pediatric, adolescent/high-risk infant, adult, and dialysis.
All formulations contain approximately 0.5 mg of aluminum (provided as amorphous aluminum hydroxyphosphate sulfate) per milliliter of vaccine.
3.Hormone - Insulin :-
Human insulin synthesized from A and B chains separately produced in Eschericdia coli from cloned synthetic genes (prepared by the Eli Lilly Research Laboratories, Indianapolis, IN) was characterized by examining its interaction with human cultured lymphocytes, human circulating erythrocytes in vitro, and isolated rat fat cells.
The binding behavior of the biosynthetic insulin with human cells was indistinguishable from that of native human or porcine insulins, with respect to affinity, association and dissociation kinetics, negative cooperativitv, and the down-regulation of lymphocyte receptors.
Similarly, the biosynthetic insulin was as potent as the native insulins in stimulating lipogenesis in isolated rat fat cells.
The basic step in recombinant DNA technology for insulin production :-
Basic about human insulin :-
1 .The insulin gene is a protein consisting of two separate chains of amino acids, an A above a B chain, that are held together with bonds. Amino acids are the basic units that build all proteins. The insulin A chain consists of 21 amino acids and the B chain has 30.
2 . Before becoming an active insulin protein, insulin is first produced as preproinsulin. This is one single long protein chain with the A and B chains not yet separated, a section in the middle linking the chains together and a signal sequence at one end telling the protein when to start secreting outside the cell. After preproinsulin, the chain evolves into proinsulin, still a single chain but without the signaling sequence. Then comes the active protein insulin, the protein without the section linking the A and B chains. At each step, the protein needs specific enzymes (proteins that carry out chemical reactions) to produce the next form of insulin.
Recombinant DNA is the general name for taking a piece of one DNA, and combining it with another strand of DNA.
Thus the name recombinant DNA is also referred as "chimera" .
By combining two or more different strands of DNA, scientists are able to create a new strand of DNA.
The most common recombinant process involves combining the DNA of two different organisms.
.Recombinant DNA formation process:-
There are three different methods by which Recombinant DNA is made. They are :-
1.Transformation,
2.Phage Introduction, and
3.Non-Bacterial Transformation.
Transformation
The first step in transformation is to select a piece of DNA to be inserted
into a vector. The second step is to cut that piece of DNA with a restriction
enzyme and then ligate the DNA insert into the vector with DNA Ligase.
The insert contains a selectable marker which allows for identification of recombinant molecules.
An antibiotic marker is often used so a host cell without a vector dies when exposed to a certain antibiotic, and the host with the vector will live because it is resistant.
The vector is inserted into a host cell, in a process called transformation. One
example of a possible host cell is E. Coli. The host cells must be specially
prepared to take up the foreign DNA.
Selectable markers can be for antibiotic resistance, color changes, or any other
characteristic which can distinguish transformed hosts from untransformed hosts.
Different vectors have different properties to make them suitable to different
applications. Some properties can include symmetrical cloning sites, size, and
high copy number.
Non-Bacterial Transformation
This is a process very similar to Transformation, which was described above. The
only difference between the two is non-bacterial does not use bacteria such as E. Coli
for the host. Ex-microinjections,biolistics etc
In microinjection, the DNA is injected directly into the nucleus of the cell being
transformed.
In biolistics, the host cells are bombarded with high velocity microprojectiles, such as particles of gold or tungsten that have been coated with DNA.
Phage Introduction
Phage introduction is the process of transfection, which is equivalent to transformation,
except a phage is used instead of bacteria. In vitro packagings of a vector is used.
This uses lambda or MI3 phages to produce phage plaques which contain recombinants.
The recombinants that are created can be identified by differences in the
recombinants and non-recombinants using various selection methods.
Application of genetic engineering in medicines :-
- In production of Better Crops (drought & heat resistance)
- In production Recombinant Vaccines (ie. Hepatitis B)
- In Prevention and cure of sickle cell anemia
- In Prevention and cure of cystic fibrosis
- In Production of clotting factors
- In Production of insulin
- In Production of recombinant pharmaceuticals
- In Plants that produce their own insecticides
- In Germ line and somatic gene therapy
- In treatment of genetic disease
Applications of rDNA (recombinant DNA) technology and genetic engineering in production of :-
- Interferons
- vaccines - hepatitis-B
- Hormones - Insulin
1.Interferons:-
Interferons are a group of signaling proteins made and released by host cells in response to the presence of several viruses. In a typical scenario, a virus-infected cell will release interferons causing nearby cells to heighten their anti-viral defenses.
A wide variety of substances can induce the production of interferon, the most obvious of which is the animal viruses.
It is matter of research whether the virus contains DNA or RNA or if it is a live or killed virus.
They do, however, vary in their ability to stimulate interferon production with the paramyxoviruses (eg Newcastle disease virus, Sendai virus) and the arboviruses (eg Semliki Forest virus) being among the most efficient.
Bacteria, phages, chlamydiae, rickettsiae, mycoplasmas, protozoa and fungi are among the other, mainly intracellular organisms, that can stimulate interferon production.
The whole organism is not necessary for the induction of interferon production; fungal extracts, endotoxins, exotoxins and various lipid and sugar fractions such as capsular polysaccharide are also effective.
The genes controlling the production of interferon a and probably that of interferon f8 are situated on chromosome 9, and that for interferon y on chromosome 12.
2.Vaccines-Hepatitis B :-
In July 1986, a recombinant hepatitis B vaccine was licensed in the United States.
This vaccine built on the knowledge that heat-inactivated serum containing hepatitis B virus (HBV) and hepatitis B surface antigen (HBsAg) was not infectious, but was immunogenic and partially protective against subsequent exposure to HBV.
HBsAg was the component that conferred protection to HBV on immunization.To produce this vaccine, the gene coding for HBsAg, or “S” gene, was inserted into an expression vector that was capable of directing the synthesis of large quantities of HBsAg in Saccharomyces cerevisiae.
The HBsAg particles expressed by and purified from the yeast cells have been demonstrated to be equivalent to the HBsAg derived from the plasma of the blood of hepatitis B chronic carriers.
The recombinant S. cerevisiae cells expressing HBsAg are grown in stirred tank fermenters. The medium used in this process is a complex fermentation medium that consists of an extract of yeast, soy peptone, dextrose, amino acids, and mineral salts.
In-process testing is conducted on the fermentation product to determine the percentage of host cells with the expression construct. At the end of the fermentation process, the HBsAg is harvested by lysing the yeast cells. It is separated by hydrophobic interaction and size-exclusion chromatography. The resulting HBsAg is assembled into 22-nm–diameter lipoprotein particles. The HBsAg is purified to greater than 99% for protein by a series of physical and chemical methods. The purified protein is treated in phosphate buffer with formaldehyde, sterile filtered, and then coprecipitated with alum (potassium aluminum sulfate) to form bulk vaccine adjuvanted with amorphous aluminum hydroxyphosphate sulfate. The vaccine contains no detectable yeast DNA but may contain not more than 1% yeast protein. In a second recombinant hepatitis B vaccine, the surface antigen expressed in S. cerevisiae cells is purified by several physiochemical steps and formulated as a suspension of the antigen absorbed on aluminum hydroxide.
The procedures used in its manufacturing result in a product that contains no more than 5% yeast protein.
No substances of human origin are used in its manufacture.
Vaccines against hepatitis B prepared from recombinant yeast cultures are noninfectious and are free of association with human blood and blood products.
Each lot of hepatitis B vaccine is tested for safety, in mice and guinea pigs, and for sterility.
QC product testing for purity and identity includes numerous chemical, biochemical, and physical assays on the final product to assure thorough characterization and lot-to-lot consistency. Quantitative immunoassays using monoclonal antibodies can be used to measure the presence of high levels of key epitopes on the yeast-derived HBsAg.
A mouse potency assay is also used to measure the immunogenicity of hepatitis B vaccines.
The effective dose capable of seroconverting 50% of the mice (ED50) is calculated.
Hepatitis B vaccines are sterile suspensions for intramuscular injection. The vaccine is supplied in four formulations: pediatric, adolescent/high-risk infant, adult, and dialysis.
All formulations contain approximately 0.5 mg of aluminum (provided as amorphous aluminum hydroxyphosphate sulfate) per milliliter of vaccine.
3.Hormone - Insulin :-
Human insulin synthesized from A and B chains separately produced in Eschericdia coli from cloned synthetic genes (prepared by the Eli Lilly Research Laboratories, Indianapolis, IN) was characterized by examining its interaction with human cultured lymphocytes, human circulating erythrocytes in vitro, and isolated rat fat cells.
The binding behavior of the biosynthetic insulin with human cells was indistinguishable from that of native human or porcine insulins, with respect to affinity, association and dissociation kinetics, negative cooperativitv, and the down-regulation of lymphocyte receptors.
Similarly, the biosynthetic insulin was as potent as the native insulins in stimulating lipogenesis in isolated rat fat cells.
The basic step in recombinant DNA technology for insulin production :-
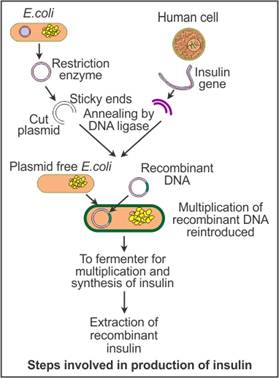
- At first suitable vector (plasmid) is isolated from E. coli and then it is cut open by restriction endonuclease enzyme.
- The gene of interest (ie. Insulin coding gene) is isolated from β-cell and inserted in opened plasmid.
- Plasmid and gene of interest are recombined together by DNA ligase enzyme
- This recombined plasmid is inserted into suitable host cell (ie E. coli) and now this recombined host cell starts producing insulin hormone.
Basic about human insulin :-
1 .The insulin gene is a protein consisting of two separate chains of amino acids, an A above a B chain, that are held together with bonds. Amino acids are the basic units that build all proteins. The insulin A chain consists of 21 amino acids and the B chain has 30.
2 . Before becoming an active insulin protein, insulin is first produced as preproinsulin. This is one single long protein chain with the A and B chains not yet separated, a section in the middle linking the chains together and a signal sequence at one end telling the protein when to start secreting outside the cell. After preproinsulin, the chain evolves into proinsulin, still a single chain but without the signaling sequence. Then comes the active protein insulin, the protein without the section linking the A and B chains. At each step, the protein needs specific enzymes (proteins that carry out chemical reactions) to produce the next form of insulin.
PCR(Polymerase chain reaction):-
- Polymerase chain reaction, or PCR, is a technique to make many copies of a specific DNA region in vitro (in a test tube rather than an organism).
- PCR depends upon thermostable DNA polymerase, Taq polymerase, and requires DNA primers designed specifically for the DNA region of interest.
- In PCR, the reaction is repeatedly cycled through a series of temperature changes, which allow many copies of the target region to be produced.
- Polymerase chain reaction (PCR) is a common laboratory technique used to make many copies (millions or billions!) of a particular region of DNA.
- PCR requires a DNA polymerase enzyme that makes new strands of DNA, using existing strands as templates. The DNA polymerase typically used in PCR is called Taq polymerase.
- Primer is a short sequence of nucleotides that provides a starting point for DNA synthesis.
- PCR primers are short pieces of single-stranded DNA, usually around 202020 nucleotides in length.
The basic steps of PCR is :-
- Denaturation (96 °\text C96°C96, °, start text, C, end text): Heat the reaction strongly to separate, or denature, the DNA strands. This provides single-stranded template for the next step.
- Annealing (555555 - 656565°\text C°C°, start text, C, end text): Cool the reaction so the primers can bind to their complementary sequences on the single-stranded template DNA.
- Extension (72 °\text C72°C72, °, start text, C, end text): Raise the reaction temperatures so Taq polymerase extends the primers, synthesizing new strands of DNA.
- Gel electrophoresis is a technique in which fragments of DNA are pulled through a gel matrix by an electric current, and it separates DNA fragments according to size.
Applications of PCR
- Using PCR, a DNA sequence can be amplified millions or billions of times, producing enough DNA copies to be analyzed using other techniques.
END

PCR(Polymerase chain reaction):-
- Polymerase chain reaction, or PCR, is a technique to make many copies of a specific DNA region in vitro (in a test tube rather than an organism).
- PCR depends upon thermostable DNA polymerase, Taq polymerase, and requires DNA primers designed specifically for the DNA region of interest.
- In PCR, the reaction is repeatedly cycled through a series of temperature changes, which allow many copies of the target region to be produced.
- Polymerase chain reaction (PCR) is a common laboratory technique used to make many copies (millions or billions!) of a particular region of DNA.
- PCR requires a DNA polymerase enzyme that makes new strands of DNA, using existing strands as templates. The DNA polymerase typically used in PCR is called Taq polymerase.
- Primer is a short sequence of nucleotides that provides a starting point for DNA synthesis.
- PCR primers are short pieces of single-stranded DNA, usually around 202020 nucleotides in length.
The basic steps of PCR is :-
- Denaturation (96 °\text C96°C96, °, start text, C, end text): Heat the reaction strongly to separate, or denature, the DNA strands. This provides single-stranded template for the next step.
- Annealing (555555 - 656565°\text C°C°, start text, C, end text): Cool the reaction so the primers can bind to their complementary sequences on the single-stranded template DNA.
- Extension (72 °\text C72°C72, °, start text, C, end text): Raise the reaction temperatures so Taq polymerase extends the primers, synthesizing new strands of DNA.
- Gel electrophoresis is a technique in which fragments of DNA are pulled through a gel matrix by an electric current, and it separates DNA fragments according to size.
Applications of PCR
- Using PCR, a DNA sequence can be amplified millions or billions of times, producing enough DNA copies to be analyzed using other techniques.
END


It's nice to have all topics at a place....nice experience
ReplyDelete